Biosciences Division
Oak Ridge National Laboratory
Telephone : (865)-241-8323
Fax : (865)-576-9939
Email : westondj@ornl.gov
Google Scholar : Profile
ORCID : 0000-0002-4794-9913
David J. Weston
Research Staff
Joint Faculty, Department of Biology, Duke University
Adjunct Professor, University of Tennessee, Ecology and Evolutionary Biology
- PhD, Clemson University, 2006
- M.S., Cornell University, 2001
- B.S., Cornell University, 1998
Research Interests:
Our research is motivated by a desire to further our understanding of the underling genetics driving plant traits critical to organismal performance and terrestrial C cycle dynamics under changing climatic conditions. Our lab takes an integrated approach using high-throughput genomic technologies, biochemistry and genetics, all placed within a physiological framework to answer our research questions. Unique to our approach is the use of hierarchical models that link systems biology statistical models (genomic predictions, metabolic & transcriptional networks) to physiological process-based models.
A guiding principle to our research is the use of model-informed experimentation to explicitly test predictions using laboratory- and field-based manipulations that ultimately feedback to improve model parameterization.
We work at multiple scales with the following projects aimed at scaling genome level inference to physiological traits and phenotypic predictions.
 CAM plant genomics and physiology: As part of a larger project (cambiodesign.org), I am working with Anne Borland (Newcastle University, UK) and Xiaohan Yang (ORNL) to link gene regulatory network models to underlying biochemical and physiological processes governing CAM (crassulacean acid metabolism) photosynthesis and plant growth. CAM is a key adaptation allowing plants to conserve water and is relatively common among desert succulents.
CAM plant genomics and physiology: As part of a larger project (cambiodesign.org), I am working with Anne Borland (Newcastle University, UK) and Xiaohan Yang (ORNL) to link gene regulatory network models to underlying biochemical and physiological processes governing CAM (crassulacean acid metabolism) photosynthesis and plant growth. CAM is a key adaptation allowing plants to conserve water and is relatively common among desert succulents.
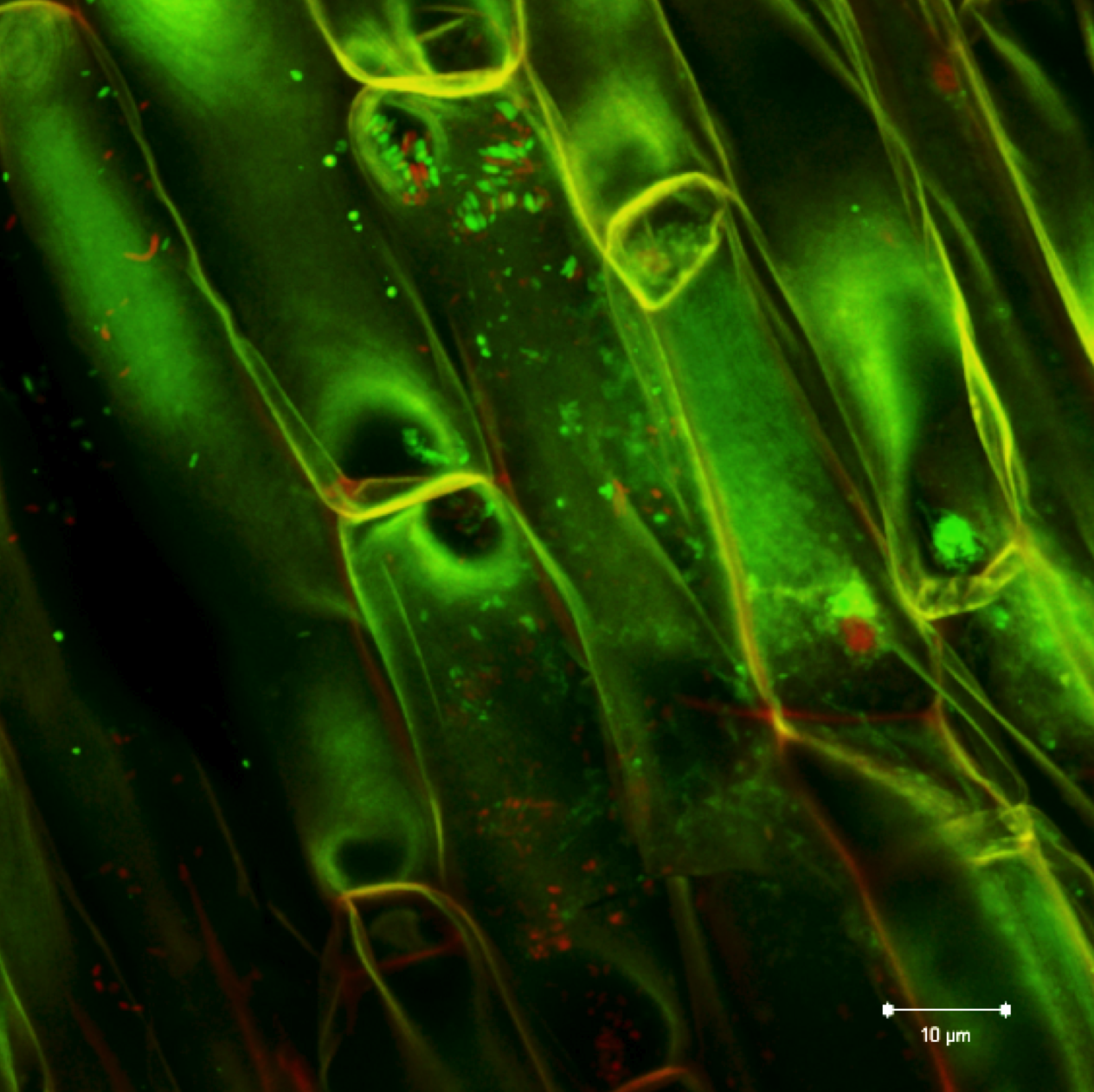 Plant and microbe interactions: As part of the Plant Microbes Interface project (pmi.ornl.gov), our primary objective is to understand how host plant genotype and environment interact with microbial members to shape community assembly and function. Populus spp. are known to associate with numerous microbes resulting in an assortment of functional interactions through known and yet to be discovered signaling and metabolic networks that drive neutral, antagonistic and mutualistic responses. Our efforts have identified systemic root-bacteria derived gene network that influences leaf metabolism, photosynthesis and whole plant fitness.
Plant and microbe interactions: As part of the Plant Microbes Interface project (pmi.ornl.gov), our primary objective is to understand how host plant genotype and environment interact with microbial members to shape community assembly and function. Populus spp. are known to associate with numerous microbes resulting in an assortment of functional interactions through known and yet to be discovered signaling and metabolic networks that drive neutral, antagonistic and mutualistic responses. Our efforts have identified systemic root-bacteria derived gene network that influences leaf metabolism, photosynthesis and whole plant fitness.
 The scientific goal of KBase (kbase.us) is to produce predictive models, reference datasets, and analytical tools and to demonstrate their utility in DOE biological research relating to bioenergy and carbon cycle. The plant component of the KBase will allow users to model genotype-to-phenotype relationships using metabolic and functional networks as well as phenotype measurements and high throughput experimental data.
The scientific goal of KBase (kbase.us) is to produce predictive models, reference datasets, and analytical tools and to demonstrate their utility in DOE biological research relating to bioenergy and carbon cycle. The plant component of the KBase will allow users to model genotype-to-phenotype relationships using metabolic and functional networks as well as phenotype measurements and high throughput experimental data.
A key challenge for accurate C cycle predictions is in understanding how multiple interacting climate change drivers influence primary productivity among diverse landscapes. I participate in two projects spanning boreal and tropic regions aimed at enhancing Earth System Model parameterization and thus prediction through plant-based trait characterizing.
 The SPRUCE project is a multi-year interaction among scientists at ORNL and the U.S. Forest Service at the Marcell Experimental Forest. Our objective within this large collaborative project is to define the response surface of gross photosynthesis to warming, tissue water content, and ambient CO2 concentration for dominant Sphagnum species. These results will be incorporated into peatland modeling efforts (with Paul Hanson), and plant community composition change among woody and herbaceous vegetation (with Rich Norby).
The SPRUCE project is a multi-year interaction among scientists at ORNL and the U.S. Forest Service at the Marcell Experimental Forest. Our objective within this large collaborative project is to define the response surface of gross photosynthesis to warming, tissue water content, and ambient CO2 concentration for dominant Sphagnum species. These results will be incorporated into peatland modeling efforts (with Paul Hanson), and plant community composition change among woody and herbaceous vegetation (with Rich Norby).
 Tropical forest research: In a recent collaboration with climate modeler Lianhong Gu (ORNL), Rich Norby (ORNL), and Klaus Winter (Smithsonian Tropical Research Institute), we are conducting gas exchange and chlorophyll fluorescence measurements to enhance predictions on tropical forest carbon budgets by improving mesophyll conductance and photosynthesis representations in climate models (e.g., CLM4).
Tropical forest research: In a recent collaboration with climate modeler Lianhong Gu (ORNL), Rich Norby (ORNL), and Klaus Winter (Smithsonian Tropical Research Institute), we are conducting gas exchange and chlorophyll fluorescence measurements to enhance predictions on tropical forest carbon budgets by improving mesophyll conductance and photosynthesis representations in climate models (e.g., CLM4).
The moss Sphagnum is a new study organism in our lab as it is a key member in peatland and arctic ecosystems that account for vast stores in terrestrial carbon. Whether these ecosystems continue to store carbon in response to changing climatic conditions remains an open question. This makes Sphagnum arguably the most important plant genus governing terrestrial carbon cycling.
Our lab is pioneering the use of Sphagnum for ecological genomics studies and together with Jon Shaw (Duke U.) and the Department of Energy (DOE) Joint Genome Institute (JGI) we are sequencing the Sphagnum fallax and Sphagnum magellanicum genomes. Furthermore, a 480 member pedigree of S. fallax has been generated with 192 members being resequenced. These genomic resources are being generated to link genomic predictions and population level metabolic networks to physiological models. To our knowledge, this marks the first plant species/population to be sequenced specifically for research related to C cycling.